
Principal Investigator
Biochemistry & Molecular Biology
zhang_adam@szbl.ac.cn
2021 - Present Shenzhen Bay Laboratory Junior Principal Investigator
2016 - 2021The Children's Hospital of Philadelphia Postdoc Fellow
2011 - 2016 University of Maryland College Park Ph.D.
2007 - 2011 Tongji University B.S
Epigenetics, genomics, 3D genomics, transcription regulation and disease, mitosis, aging.
• Established an in-vitro induction and differentiation system for vascular smooth muscle cells from premature aging HGPS patients.
• Revealed the molecular mechanism of HGPS smooth muscle cell death, providing noval therapeutic potential for the treatment of HGPS patients.
• Revealed the process of chromatin 3D structural reconfiguration during the mitosis to G1 phase transition; uncovered the dynamic process of post-mitotic chromatin recruitment of the architectural protein CTCF and cohesin ring complex; provides noval supports for the regulation role of loop extrusion model in post-mitotic chromatin 3D structural reconfiguration.
• Uncovered the relationship between chromatin 3D structural reconfiguration and transcription reactivation after mitosis.
• Uncovered the role of architecture protein CTCF in the reconfiguration of chromatin 3D structure after mitosis.
Has been publishing in prestigious journals including Nature, PNAS, Aging Cell etc.
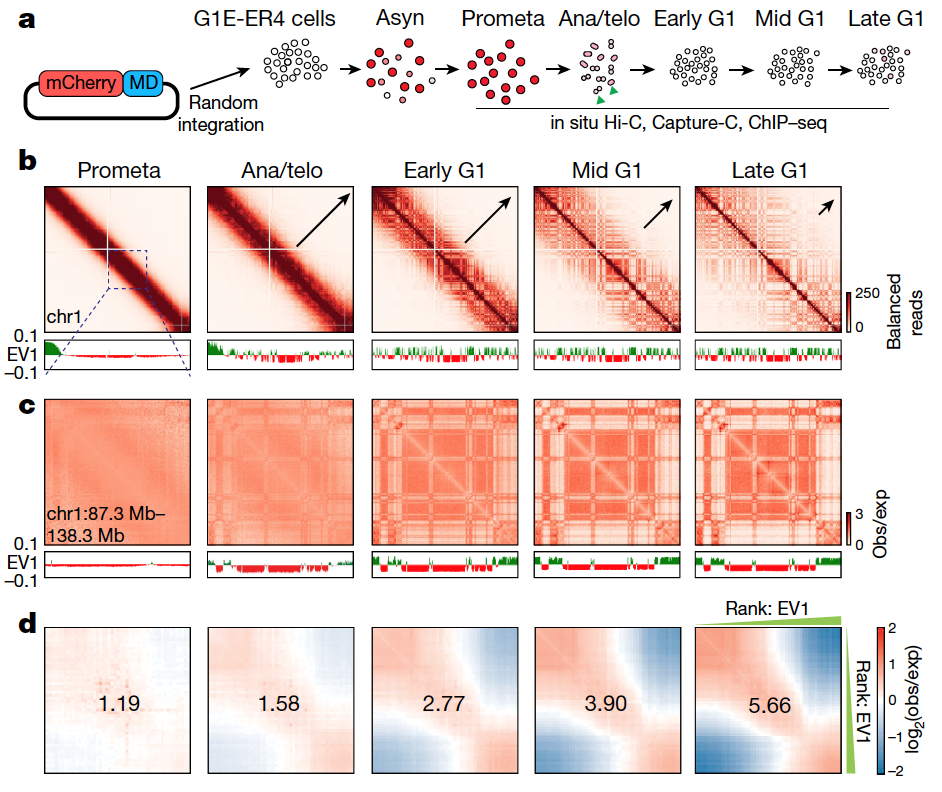
Reestablishment of chromatin A/B compartments during the mitosis to G1 phase transition
Preprints
1. Aboreden, N.,…Zhang H* & Blobel, G*. Cis-regulatory chromatin contacts form de novo in the absence of loop extrusion. BioRxiv, 2025.01. 12.632634
Primary research publications
1. Li T., Wang G.,…Zhang H*. & Yang J*. Estrogen receptor 1 signaling in hepatic stellate cells designates resistance to liver fibrosis. Cell Discovery, (2025). https://doi.org/10.1038/s41421-025-00783-3
2. Zhao H., Shu L.,…Zhang H*. Extensive mutual influences of SMC complexes shape 3D genome folding. Nature, (2025). https://doi.org/10.1038/s41586-025-08638-3
3. Zhao H.,…Zhang H*. Genome folding principles uncovered in condensin-deficient mitotic chromosomes. Nature Genetics, 56(6):1213-1224 (2024).
4. Zhang H*. et al. CTCF and transcription influence chromatin structure re-configuration after mitosis. Nat Commun 12, 5157 (2021).
5. Zhang H. et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158-162 (2019).
6. Zhang H. et al. Loss of H3K9me3 Correlates with ATM Activation and Histone H2AX Phosphorylation Deficiencies in Hutchinson-Gilford Progeria Syndrome. PLoS One 11, e0167454 (2016).
7. Zhang H., Xiong, Z.M. & Cao, K. Mechanisms controlling the smooth muscle cell death in progeria via down-regulation of poly(ADP-ribose) polymerase 1. Proc Natl Acad Sci USA 111, E2261-70 (2014)
Review publications
1. Zhang H*. & Blobel, G.A. Genome folding dynamics during the M-to-G1-phase transition. Curr Opin Genet Dev 80, 102036 (2023).
2. Zhang H and Cao K. Mechanisms of Genome Instability in Hutchinson-Gilford Progeria. Frontiers in Biology, DOI 10.1007/s11515-016-1435-x (2016)
3. Zhang H., Kieckhaefer, J.E. & Cao, K. Mouse models of laminopathies. Aging Cell 12, 2-10 (2013).
Other publications
1. Aboreden Nicholas,…, Zhang Haoyue, Hansen Anders, Blobel Gerd*. LDB1 establishes multi-enhancer networks to regulate gene expression. Molecular Cell (2025).
2. Lam Jessica,…, Zhang Haoyue, Blobel Gerd*. YY1-controlled regulatory connectivity and transcription are influenced by the cell cycle. Nature Genetics 56, 1938-1952, doi:10.1038/s41588-024-01871-y (2024).
3. Luan J, Vermunt MW, Syrett CM, Cote A, Tome JM, Zhang H, et al. CTCF blocks antisense transcription initiation at divergent promoters. Nature structural & molecular biology. 2022;29(11):1136-44.
4. Linares-Saldana R, Kim W, Bolar NA, Zhang H, Koch-Bojalad BA, Yoon S, et al. BRD4 orchestrates genome folding to promote neural crest differentiation. Nat Genet. 2021;53(10):1480-92.
5. Luan J, Xiang G, Gomez-Garcia PA, Tome JM, Zhang Z, Vermunt MW, Zhang H, et al. Distinct properties and functions of CTCF revealed by a rapidly inducible degron system. Cell reports. 2021;34(8):108783.
6. Zhang D, Huang P, Sharma M, Keller CA, Giardine B, Zhang H, et al. Alteration of genome folding via contact domain boundary insertion. Nat Genet. 2020;52(10):1076-87.
7. Wang K, Wu D, Zhang H, Das A, Basu M, Malin J, et al. Comprehensive map of age-associated splicing changes across human tissues and their contributions to age-associated diseases. Sci Rep. 2018;8(1):10929.
8. Atchison L, Zhang H, Cao K, and Truskey GA. A Tissue Engineered Blood Vessel Model of Hutchinson-Gilford Progeria Syndrome Using Human iPSC-derived Smooth Muscle Cells. Sci Rep. 2017;7(1):8168.
9. Tariq Z, Zhang H, Chia-Liu A, Shen Y, Gete Y, Xiong ZM, et al. Lamin A and microtubules collaborate to maintain nuclear morphology. Nucleus (Austin, Tex). 2017;8(4):433-46.
10. Wu D, Yates PA, Zhang H, and Cao K. Comparing lamin proteins post-translational relative stability using a 2A peptide-based system reveals elevated resistance of progerin to cellular degradation. Nucleus (Austin, Tex). 2016;7(6):585-96.
11. Xiong ZM, Choi JY, Wang K, Zhang H, Tariq Z, Wu D, et al. Methylene blue alleviates nuclear and mitochondrial abnormalities in progeria. Aging cell. 2015.
12. McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome research. 2013;23(2):260-9.