
Principal Investigator
Genetics
sunkun@szbl.ac.cn
2019-PresentShenzhen Bay Laboratory Junior Principal Investigator
2017 - 2019The Chinese University of Hong Kong Research Assistant Professor
2014 - 2017 The Chinese University of Hong Kong Postdoctoral Fellowship
2006 - 2010 University of Science and Technology of China B.S.
- Cancer liquid biopsy
- Bioinformatics/Computational biology
- Long noncoding RNA
- Big data mining, Artificial Intelligence (AI), and data visualization
Dr. Kun Sun has been engaged in bioinformatics and blood-based liquid biopsy in cancer diagnosis for more than 10 years. He has published 46 papers, 27 of which Dr. Sun serves as the (co-)first and/or (co-)corresponding author, including papers published on Cancer Discovery, Genome Research, PNAS, EMBO Journal, etc. The total citation of Dr. Sun’s publications is more than 2,600.
Dr. Sun has developed a method known as “plasma DNA tissue mapping” for deconvoluting plasma cell-free DNA compositions for tissue-of-origin predictions of the tumors in cancer patients (Sun et al. PNAS 2015), which work has far-reaching influence in the cancer liquid biopsy field. By investigating the cleavage mechanism of cell-free DNA, Dr. Sun identified various tissue-specific cfDNA fragmentation signatures and demonstrated their potential as biomarkers in noninvasive prenatal diagnosis and cancer diagnosis (Chan et al. PNAS 2016; Sun et al. PNAS 2018; Jiang et al. PNAS 2018, Sun et al. Genome Research 2019). With other researchers, Dr. Sun discovered that DNASE1L3 is an important nuclease for cell-free DNA formation (Lee et al. PNAS 2019) which directly affects the cell-free DNA end motif pattern and provides a new class of biomarkers for cancer diagnosis (Jiang et al. Cancer Discovery 2020).
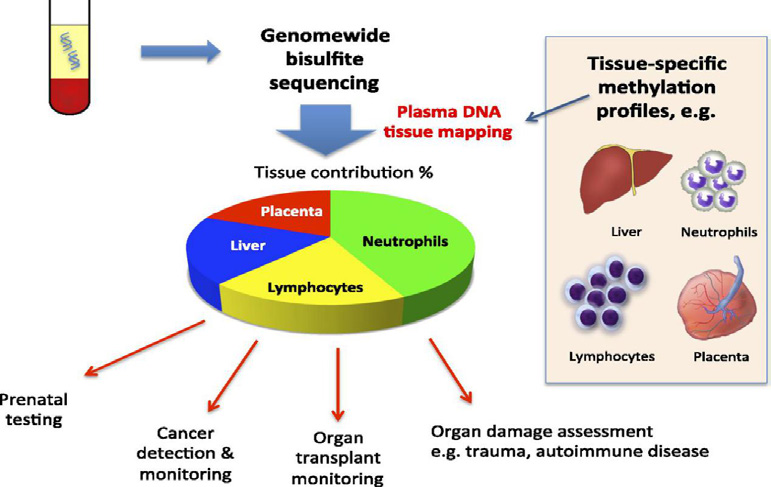
Fig. 1: Tissue-of-origin tracing of plasma DNA
Dr. Sun has also worked in long non-coding RNA (lncRNA) fields for a long time. With other researchers, he reported that transcription factors could directly participate in the regulation of lncRNAs (Lu et al. Embo Journal 2013) and lncRNAs could regulate the binding transcription factors to DNA (Zhou et al. Nature Communications 2015), revealing new mechanisms in the interactions between lncRNAs and transcription factors. In addition, Dr. Sun has developed several bioinformatics tools for lncRNA identification (Sun et al BMC Genomics 2013; Sun et al. Briefings in Bioinformatics 2020).

Fig. 2. New mechanisms in the interactions between lncRNAs and transcription factors.
An Y, Zhao X, Zhang Z, Xia Z, Yang M, Ma L, Zhao Y, Xu G, Du S, Wu X, Zhang S, Hong X, Jin X*, Sun K*. DNA methylation analysis explores the molecular basis of plasma cell-free DNA fragmentation. Nat Commun 2023 Jan 18; 14(1):287
Jiang P#, Sun K#, Peng W#, Cheng SH, Ni M, Yeung PC, Heung MMS, Xie T, Shang H, Zhou Z, Chan RWY, Wong J, Wong VWS, Poon LC, Leung TY, Lam WKJ, Chan JYK, Chan HLY, Chan KCA, Chiu RWK, Lo YMD. Plasma DNA end motif profiling as a fragmentomic marker in cancer, pregnancy and transplantation. Cancer Discov 2020 May; 10(5):664-673.
Sun K*, Jiang P, Cheng SH, Cheng THT, Wong J, Wong VWS, Ng SSM, Ma BBY, Leung TY, Chan SL, Mok TSK, Lai PBS, Chan HLY, Sun H, Chan KCA, Chiu RWK, Lo YMD*. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res 2019 Mar; 29(3):418-427.
Sun K, Jiang P, Wong AIC, Cheng YKY, Cheng SH, Zhang H, Chan KCA, Leung TY, Chiu RWK, Lo YMD. Size-tagged preferred ends in maternal plasma DNA shed light on production mechanism and show utility in noninvasive prenatal testing. Proc Natl Acad Sci U S A 2018 May 29; 115(22):E5106-E5114
Sun K#, Jiang P#, Chan KCA#, Wong J, Cheng YK, Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, Chan RW, Tong YK, Ng SS, Mong RSM, Hui DS, Leung TN, Leung TY, Lai PBS, Chiu RWK, Lo YMD. Plasma DNA tissue mapping by genomewide methylation sequencing for noninvasive prenatal, cancer and transplantation assessments. Proc Natl Acad Sci U S A 2015 Oct 6; 112(40):E5503-12.
Sun K*, Wang H, Sun H*. NAMS webserver: coding potential assessment and functional annotation of plant transcripts. Brief Bioinform 2021 May 20; 22(3):bbaa200.
Sun K*, Li L, Ma L, Zhao Y, Deng L, Wang H, Sun H. Msuite: a high-performance and versatile DNA methylation data-analysis toolkit. Patterns (N Y) 2020 Oct 15; 1(8):100127.
Zhou L#, Sun K#, Zhao Y#, Zhang S, Wang X, Li Y, Lu L, Chen X, Chen F, Bao X, Zhu X, Wang L, Tang LY, Esteban MA, Wang R, Jauch R, Sun H, Wang H. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun 2015 Dec 11; 6:10026.
Lu L#, Sun K#, Chen X, Zhao Y, Wang L, Zhou L, Sun H, Wang H. Genome-wide survey by ChIP-seq reveals YY1 regulation of lincRNAs in skeletal myogenesis. EMBO J 2013 Oct 2; 32(19):2575-88.