
Senior Principal Investigator
Biochemistry & Molecular Biology
zhouyq@szbl.ac.cn
2021 - PresentShenzhen Bay Laboratory, Shenzhen,China Senior Principal Investigator
2013~2021Griffith University, QLD, Australia Professor
2006-2013Indiana University Purdue University at Indianapolis, Indiana,USA Tenured Full Professor
2004-2006 State University of New York at Buffalo,USA Tenured Associate Professor
2000-2004 State University of New York at Buffalo,USA Assistant Professor
1995-2000 Harvard University,MA,USA Research Associate
1994-1995North Carolina State University,NC,USA Postdoctoral Fellow
1994-1994 State University of New York at Stony Brook,NY,USA Postdoctoral Fellow
1990-1993Applied P & Ch Laboratory,CA,USA Scientist and Director
The research group led by Dr. Zhou primarily focuses on fundamental studies exploring the relationships between sequences, structures, and functions of RNA and proteins, as well as applied research in design, delivery, and drug development of these biological macromolecules. A distinctive feature of the group is the integration of dry and wet lab approaches—combining computational structural bioinformatics, AI-powered deep learning with modern high-throughput and automated directed-evolution biotechnologies—to achieve profound insights into the sequence-structure-function relationships. This research enables the multi-faceted application of biological macromolecules, including targeted drug design and delivery for precision medicine, as well as detection of personalized biomarkers. Current specific projects include, but are not limited to:
· Protein structure and function prediction in the post-AlphaFold era
· Protein design and directed evolution
· RNA structure prediction and RNA language models
· Targeted RNA delivery systems
· Pioneered prediction of continuous backbone dihedral angles using shallow and deep learning, enabling end-to-end protein structure prediction (Fig. 1). This development directly underpinned Nobel-prize-winning technique AlphaFold 2 for high-accuracy structure prediction.
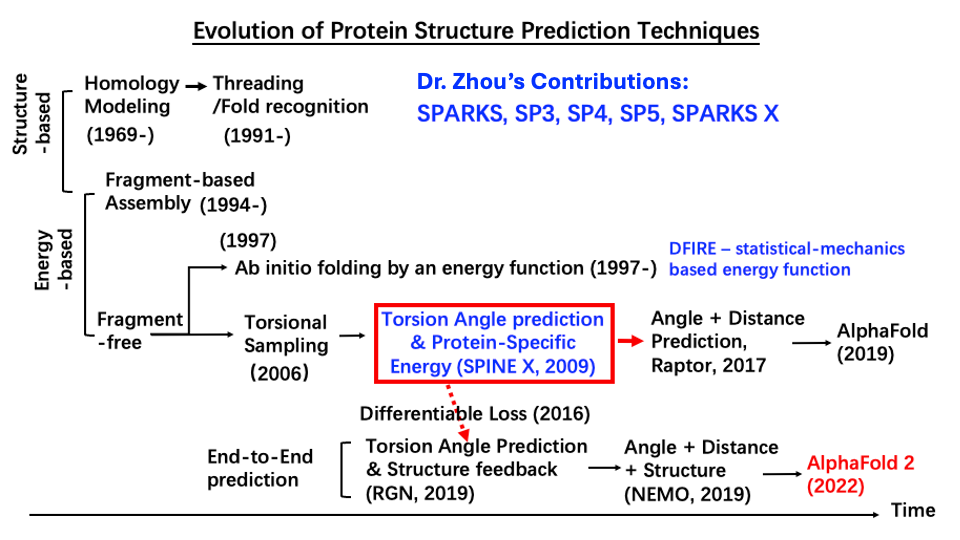
Fig. 1
· First AI-driven methods for protein sequence design, achieving 30–34% sequence recovery (Fig. 2). Recognized as the inception of machine learning in protein design ([Ovchinnikov and Huang, Curr. Opin. Chem. Biol. 2021]), this paradigm shift (away from energy-based approaches) now dominates the field, enabling revolutionary advances in therapeutic protein & industrial enzyme design.
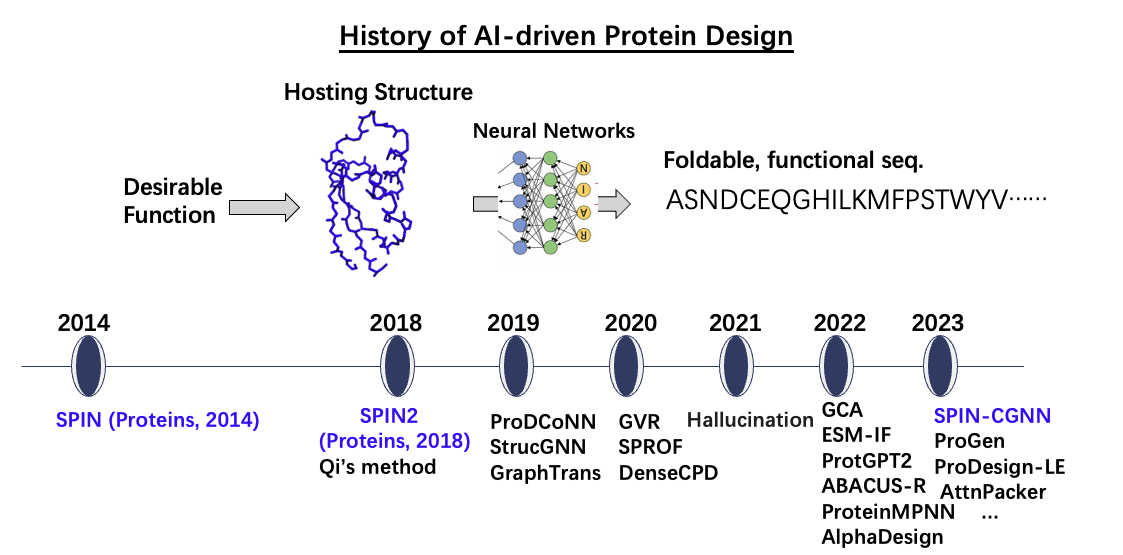
Fig. 2
· RNA-BRiQ: Developed a novel statistical potential enabling atomic-level RNA refinement. Powered AIChemy-RNA2 to win #1 in CASP15 (2022) for RNA structure prediction.
· Top 2% Scientist Worldwide in Stanford/Elsevier’s career and annual rankings
· 20,000+ citations & an H-index of 79 (Google Scholar)
· #1 Performances in International Competitions: CASP6 Template-based protein structure prediction (2004), CAGI Blind prediction of cell proliferation rate (2013), CAID Prediction of protein unstructured regions (2019), CASP15 RNA structure prediction (2022)
· Editorial Board, Nucleic Acids Research
1. Y. Zhou, T. Litfin, and J. Zhan, 3=1+2: How the Divide Conquered de Novo Protein Structure Prediction and What’s Next? National Science Review, 10:nwad259 (2023).
2. P. Xiong, R. Wu, J. Zhan, and Y. Zhou, “Pairing a high-resolution statistical potential with a nucleobase-centric sampling algorithm for improving RNA model refinement.”, Nature Communications, 12,2777 (2021).
3. J. Singh, J. Hanson, K. Paliwal, and Y. Zhou, RNA secondary structure prediction using an ensemble of two-dimensional deep neural networks and transfer learning, Nature Communications 10, 5407 (2019).
4. Z. Zhang, P. Xiong, T. Zhang, J. Wang, J. Zhan, and Y. Zhou, Accurate inference of the full base-pairing structure of RNA by deep mutational scanning and covariation-induced deviation of activity, Nucleic Acids Research, 48:1451-1465 (2020).
5. J. Zhan, H. Jia, E. A. Semchenko, Y. Bian, A. M. Zhou, Z. Li, Y. Yang, J. Wang, S. Sarkar, M. Totsika, H. Blanchard, F. E.-C. Jen, Q. Ye, T. Haselhorst, M. P. Jennings, K. L. Seib, and Y. Zhou, Self-derived structure-disrupting peptides targeting methionine aminopeptidase in pathogenic bacteria; a new strategy to generate antimicrobial peptides, FASEB J. , 33: 2095–2104 (2019).
6. Z. Li, Y. Yang, J. Zhan, L. Dai and Y. Zhou, Energy Functions in De Novo Protein Design: Current Challenges and Future Prospects, Ann. Rev. Biophysics 42, 315-335 (2013).